Accordion Content
-
Plastid transformation has been inefficient in Arabidopsis thaliana due to a natural tolerance of Arabidopsis to spectinomycin, the selective agent employed to enrich transformed plastid genomes. Tolerance to spectinomycin has been linked to a duplication of the ACCase biosynthetic pathway in chloroplasts. We have shown that plastid transformation is 100-fold more efficient in Arabidopsis lines defective in the plastid-targeted ACC2 nuclear gene (Yu et al. Plant Physiol. 175: 186-193, 2017). This information has been obtained in the the Col-0 ecotype that is recalcitrant to plant regeneration. We now report ACC2 defective lines in the RLD and Ws ecotypes, which readily regenerate plants from cultured cells. ACC2 knockouts were obtained using CRISPR/Cas9 genome editing tools. The spectinomycin hypersensitive phenotype is characterized by the lack of shoot apex when germinated on a selective medium. This phenotype has been confirmed in both accessions, indicating that deletion of the ACC2 gene is generally applicable to obtain spectinomycin hypersensitive plants in all species in which duplication of the ACCase pathway has been conserved. Testing plastid transformation efficiency in the ACC2 knockout lines confirmed 100-fold elevated frequency as compared to the wild-type. Testing was accelerated by the newly developed SPEED transformation protocol (Yu et al., Plant Physiol. 181: 394-398, 2020). The research is supported by NSF award MCB-1406971.
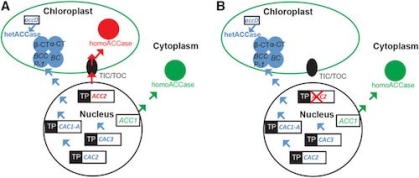
Elimination of ACC2 function makes plastid transformation efficient in Arabidopsis. Based on Fig. 1 in Yu et al. Plant Physiol. 175: 186-193, 2017. -
The current bottleneck of plastid transformation in Arabidopsis is the difficulty of obtaining fertile plants from transplastomic tissue culture cells. Tissue culture limitations in Arabidopsis nuclear gene transformation were overcome by using Agrobacterium to directly transform the female gametocyte, and identification of nuclear transgenic events by germinating the resulting seedlings on a selective medium. Our goal is to re-engineer Agrobacterium for T-DNA delivery to chloroplasts to directly transform the plastids in the female gametocyte. T-DNA export from Agrobacterium to plant cells occurs by the type 4 protein secretion machinery. Recently, we obtained proof of concept that proteins can be directly exported from Agrobacterium to chloroplasts. The protein of our choice was the phiC31 phage site-specific integrase (Int), because visitation of the recombinase to chloroplasts created a permanent footprint. We are now working on re-targeting the proteins involved in T-DNA transfer. Side-stepping the tissue culture process will eliminate the need for specialized expertise to practice plastid transformation in Arabidopsis. Therefore, this research will lead to widespread applications of Arabidopsis plastid genome engineering which, combined with the available extensive genomic resources, will have a major impact on basic science and applications in biotechnology. This research is supported by NSF Award IOS-2037155.
-
The laboratory has a long tradition in the expression of recombinant proteins in chloroplasts. Most recent is development of dicistronic operons as a novel marker system for chloroplast engineering that can be used as building blocks for plant synthetic biology. The identification of transplastomic clones is based on selection for antibiotic resistance encoded in the first open reading frame (ORF) and accumulation of the reporter gene product in tobacco chloroplasts encoded in the second ORF. The antibiotic resistance gene may encode spectinomycin or kanamycin resistance based on the expression of aadA or neo genes, respectively. The reporter gene used in the study is the green fluorescent protein (GFP). The mRNA level depends on the 5’ UTR of the first ORF. The protein output depends on the strengths of the ribosome binding, and is proportional with the level of translatable mRNA. Because the dicistronic mRNA is not processed, we could show that protein output from the second ORF is independent from the first ORF (Figure 1). High-level GFP accumulation from the second ORF facilitates identification of transplastomic events under UV light. Expression of multiple proteins from an unprocessed mRNA is an experimental design that enables predictable protein output from polycistronic mRNAs, expanding the toolkit of plant synthetic biology. These data have been described in a recent publication (Yu et al., 2020 Plant Journal doi: 10.1111/tpj.14864).
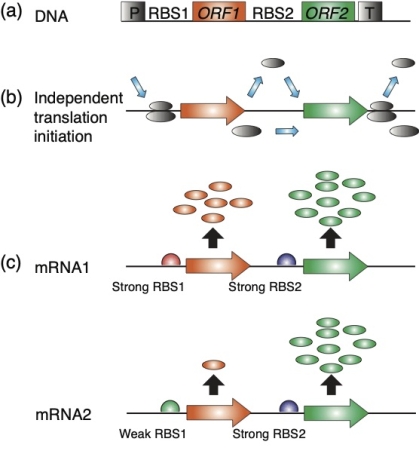
Figure 1. No translational coupling between the first and second ORF in the chloroplast dicistronic markers. Based on Figure 6 in Yu et al. Plant J. doi: 10.1111/tpj.14864
(a) Schematic map of dicistronic operons. ORF1 encodes a marker gene, the second ORF gfp. The operon has one promoter (P) and one 3’-UTR (T) for the stabilization of the mRNA.
(b) Translation initiates independently from the first and second ORFs. Shown are the small and large ribosomal subunits entering the mRNA at the ribosome entry site, and dissociating when translation of the 1st ORF is completed. Small ribosomal subunits initiate translation independently of the 2nd ORF.
(c) Protein accumulation from the first ORF has no significant impact on protein accumulation from the 2nd ORF. Protein output from the 1st ORF can be high (pMRR20) or low (pMRR21), the protein output from the 2nd ORF is always high -
Engaging undergraduate students in research is part of the broader impact of research activity supported by the National Science Foundation. We integrate research and education by training undergraduates to facilitate full participation of women and underrepresented minorities in STEM fields. We also host students from Farmingdale State College, a Primarily Undergraduate Institution, to expose the Farmingdale students to the research University environment. The students come through our collaborator, Associate Professor Kerry A. Lutz, who is Co-PI on the NSF Grants MCB-1506971 and IOS-2037155 supporting research experience for undergraduates. The name of undergraduate coauthors in the list of publications is in bold face.