Accordion Content
-
The conserved KMN network is required for KT-MT attachments in vivo and is composed of three groups of proteins: SPC105/KNL1, the MIS12 complex and the NDC80 complex. NDC80 is required for end-on MT attachments, which are important for moving the chromosomes towards the poles at anaphase. We have identified several activities that depend on SPC105R, kinetochore assembly (NDC80 recruitment), lateral microtubule attachments, and sister centromere fusion. The latter is important for coorientation, the mechanism that promotes attachment of sister kinetochores in meiosis I to microtubules from the same pole.
To study the function of SPC105R, we use tissue-specific RNAi to deplete the protein in oocytes. To develop a system to study the functions of SPC105R in oocytes with mutants, we generated an Spc105R RNAi-resistant transgene. This transgene rescues all Spc105R RNAi phenotypes. Using this construct, we are generating RNAi-resistant transgenes with deletions of each SPC105R domain proposed to recruit proteins with specific functions (Figure 1). We are particularly interested in separation-of-function mutants that identify the region(s) required for lateral attachments and sister centromere fusion. Surprisingly, the large central domain and the N-terminal MT-binding domains are not essential for meiosis. However, a transgene expressing only the C-terminal domain appears to be sufficient to localize to the centromeres, recruit other kinetochore proteins, and most surprising, recruits cohesin protection proteins such as MEI-S332/Shugoshin. These studies are continuing with the generation and characterization of additional mutations.
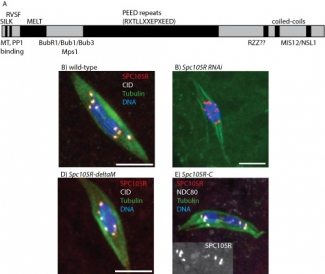
Mutational analysis of Spc105R 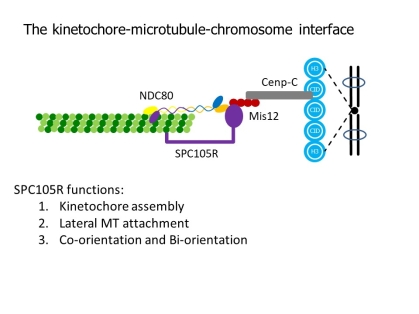
kt parts -
Prior to meiosis, the DNA replicates. Cohesin is the protein complex that holds these sister chromatids together until meiosis I and II. Cohesin regulation is complex because it must be protected from destruction prior to these stages, and then released in stages. First the chromosomes arms during meiosis I, and then the centromeres and the pericentromeric regions in meiosis II. regulate sister centromere fusion. The analysis of kinetochore proteins SPC105R/KNL1 and Protein Phosphatase 1 (PP1-87B) has shown that two independent mechanisms maintain sister centromere cohesion in Drosophila oocytes. Maintenance of sister centromere cohesion by SPC105R involves protection against Separase, the enzyme that’s destroys cohesin proteins. In contrast, maintenance of sister centromere cohesion by PP1-87B does not involve Separase. Instead, PP1-87B maintains sister centromere cohesion by inhibiting stable KT-MT attachments. In a recent paper describing this work, we propose two mechanisms regulate co-orientation in Drosophila oocytes. First, SPC105R protects cohesins at sister centromeres to maintain fusion. Second, separation of sister centromeres in meiosis II occurs without degrading cohesins, depends on end-on microtubule attachments, and during meiosis I is prevented by PP1-87B mediated destabilization of these attachments.
SPC105R maintains sister centromere cohesion during meiosis I by recruiting Protein Phosphatase 2A (PP2A), which modifies cohesins so they cannot be destroyed. What regulates stepwise removal of cohesion in the pericentromeric regions and the chromosome arms is not kown. This analysis is complicated by the fact that Drosophila have two PP2A subunits, WDB and WRD, that are partially redundant. In oocytes lacking SPC105R, WDB does not localize to the centromeres. Wdb RNAi oocytes, however, show a mild defect in cohesion, displaying occasionally showing separated centromeres and precocious anaphase. When both WDB and WRD are depleted by RNAi, we observed a complete failure to maintain sister chromatid and arm cohesion in metaphase I oocytes (Figure 2). We are currently investigating how PP2A is recruited to the pericentromeric regions and chromosome arms. Based on published work in mitotic cells, we are determining if Dalmatian, the Drosophila homolog of cohesin regulator Sororin, recruits PP2A to the chromosomes in addition to MEI-S332/Shugoshin. Our preliminary data shows that Dalmatian is localized to the meiotic chromosomes during early prophase. It may protect cohesion during the long prophase arrest experienced by oocytes, and during meiosis I may be redundant with MEI-S332/Shugoshin.
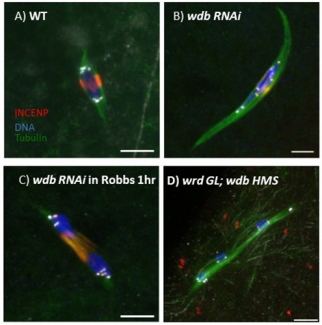
Phosphatases -
A key player in Drosophila meiosis is the four-protein chromosomal passenger complex (CPC). INCENP (Figure 2A) targets the complex to specific structures and recruits Aurora B, a kinase and the catalytic component of the CPC. In females expressing RNAi to Incenp or Aurora B, no meiotic spindle forms, showing that the CPC is required for chromosomes mediated spindle formation in oocytes.
To understand how the CPC is recruited to the chromosomes and promotes spindle assembly, we have generated mutants of Incenp that alter the localization pattern of the CPC. These mutated transgenes contain silent changes in the sequence that makes them RNAi resistant. In females expressing an RNAi resistant form of the wild-type gene, the meiotic spindle forms normally. When mutant Incenp transgenes that targeted the CPC to the centromeres or kinetochores are used, only kinetochore microtubules form, and there is no central spindle. This result suggests that targeting the CPC to the centromeres is sufficient to form a kinetochore but is not sufficient to promote bipolar spindle assembly. Several lines of evidence suggest that the key to CPC-mediated spindle assembly is heterochromatin protein 1 (HP1). First, in the absence of Aurora B, the rest of the CPC components (INCENP, Borealin, Survivin) colocalize with HP1 on the chromosomes. Second, mutants of Incenp or CPC subunit Borealin that disrupt HP1 interactions have meiotic spindle assembly defects. These experiments have shown that an interaction between HP1 and Borealin is required for the initial recruitment of the CPC to the chromosomes and the initiation of spindle assembly. Third, after spindle assembly, HP1 is mostly localized to the spindle. We propose a model for how HP1 orchestrates chromosome-mediated spindle assembly. HP1 is initially recruited to the heterochromatin by H3K9me2/3, where it recruits the CPC to promote spindle assembly. Aurora B then phosphorylates H3S10, which is known to disrupt HP1 localization. Therefore, the activity of Aurora B my cause the dissociation of HP1 from the chromatin, which may then force the CPC to move from the chromatin to the microtubules. Once on the spindle, the CPC promotes homologous chromosomes biorientation.
To identify the phosphatases that regulate Aurora B activity, we used a small molecule inhibitor of Aurora B, Binuclein 2 (BN2). When oocytes are treated with this drug, they lost their spindle microtubules. Thus, continuous Aurora B activity is required to preserve the spindle during meiosis I and spindle dynamics may be regulated by a phosphatase. When Aurora B was inhibited in PP2A RNAi oocytes, the spindle was maintained, demonstrating that PP2A antagonizes the Aurora B spindle maintenance function. We have further shown that the CPC negatively regulates KLP10A, a kinesin 13 that depolymerizes microtubules. Thus, we have found that PP2A opposes CPC functions, probably by dephosphorylating Aurora B spindle targets.
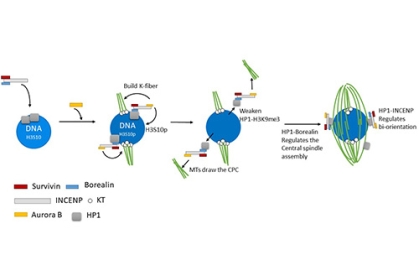
CPC loading 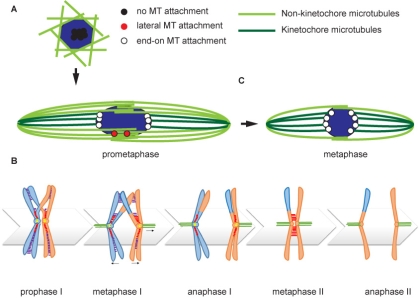
bi-orientation model -
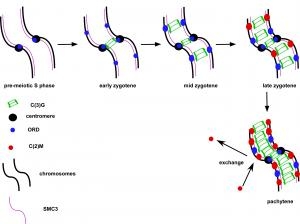
Pairs of meiotic chromosomes, or homologs, are brought together in an elaborate pairing process which culminates with synapsis, where bivalents are held together along their entire length by the synaptonemal complex (SC). Formation of the SC between homologs is essential for crossing over and chromosome segregation but how the homologs are paired and SC assembly initiates is poorly understood. We investigated the requirements for SC assembly in Drosophila oocytes and found that there are three temporally and genetically distinct stages of synapsis initiation (Figure 1). First, early zygotene cells have one or two patches of SC that colocalizes with a cluster of centromeres, suggesting synapsis initiates first at the centromeres. Second, oocytes at mid-zygotene contain the centromere SC plus several euchromatic sites. The centromeric and first euchromatic SC initiation sites depend on the cohesin protein ORD. Third, late zygotene contains many more sites of SC initiation and these depend on the Kleisin-like protein C(2)M. Surprisingly, the synapsis initiation events in late zygotene are independent of the earlier mid-zygotene events but all synapsis depend on the cohesin subunit SMC3. Our results show that cohesin proteins have an important role in SC initiation. Based on the observation that ORD and SMC3 are enriched at centromeres and promote their clustering and synapsis, we suggest that the enrichment of cohesin proteins at specific sites promotes homolog interactions and the initiation of euchromatic SC assembly. ORD is also required for most crossing over, suggesting that SC initiation at mid-zygotene may also trigger the formation of DSBs that will be repaired as crossovers.
cohesion proteins 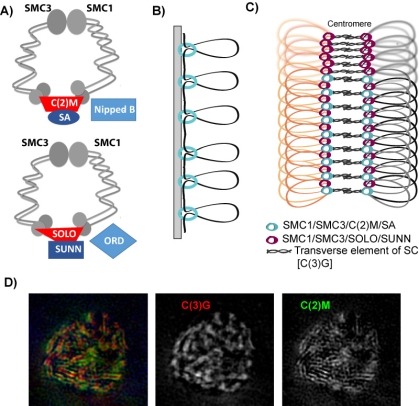
cohesion-SC model -
Meiotic recombination occurs during prophase I when homologous chromosomes are synapsed along their entire length. Synapsis is defined as the close and stable association of homologous chromosomes through a proteinaceous structure called the synaptonemal complex (SC). In most organisms, this complex is composed of two main parts: lateral elements that attach along the axis of each homologous chromosome and transverse elements that span the central region of the SC and function to tether the homologs [1,2]. At the leptotene/zygotene stages of meiotic prophase, these structural proteins begin to load onto the chromosome axes, and are completely assembled at pachytene, when homologous chromosomes are synapsed along their entire length. Recombination between the homologous chromosomes initiates with DNA double-strand breaks (DSBs) that are repaired as either crossovers or noncrossovers [3–5]. Crossovers establish chromatin linkages called chiasmata, which, along with sister chromatid cohesion, hold homologs together after recombination has been completed and chromosomes have dissociated their SC proteins. Chiasmata help orient the homologous chromosomes on the metaphase I spindle and ensure their proper segregation at anaphase I. The failure to establish a crossover/chiasma can result in the nondisjunction of homologs and lead to aneuploid gametes.
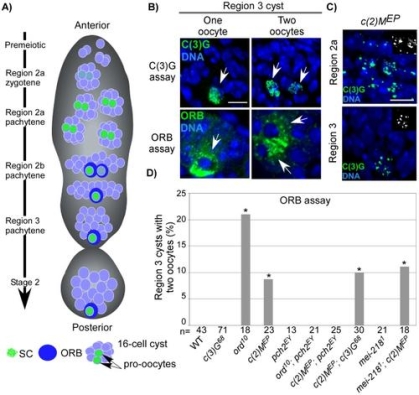
Pachytene delays in axis-defective mutants.